
On May 11, Ma Yingxin's research group of the Institute of Synthetic Biology of Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, and Dr. Yin Wen of Hubei University, jointly published a supplementary cover paper in Analytical Chemistry under the title of “Fluorescence and Colorimetric Analysis of African Swine Fever Virus Based on RPA-Assisted CRISPR/Cas12a Strategy”. In this paper, a colorimetric - fluorescence dual-mode detection strategy of African swine fever virus (ASFV) based on the RPA-CRISPR/Cas12a system was constructed for the first time. By constructing a new magnetic bead - enzyme reporting system, the precise detection of ASFV gene by colorimetric - fluorescence dual-mode was realized based on RPA-CRISPR/Cas12a technology. This method can detect single copy virus genomes, can be used for portable visual diagnosis, and has shown good detection performance in clinical sample applications, providing a powerful tool for rapid and precise diagnosis of virus infection.
Screenshot of the published paper
Link of the paper: https://pubs.acs.org/doi/pdf/10.1021/acs.analchem.3c01033
African swine fever is an acute, hemorrhagic, and highly contagious swine disease caused by the African swine fever virus. So far, there is still no widely effective vaccine or treatment for responding to African swine fever, and the outbreak is usually effectively controlled by rapid diagnosis and culling of infected swine. At present, the diagnosis of African swine fever mainly relies on PCR technology, which cannot meet the needs of on-site diagnosis since it relies on instruments and equipment and professional operators and can only be carried out in the laboratory. Isothermal amplification techniques, such as recombinase polymerase amplification (RPA) and rolling circle amplification (RCA), have the advantages of high efficiency and simplicity, but their sensitivity is limited, so that they are difficult to be used alone for molecular diagnosis. By combining them with the CRISPR/Cas system, the sensitivity is significantly improved, which can increase the accuracy of molecular diagnosis. However, the commonly used single mode fluorescence detection method or colorimetric detection method is susceptible to environmental interference, resulting in inaccurate detection results.
In this study, a novel reporting system combining RPA, CRISPR/Cas12a, and magnetic bead - enzyme was developed for rapid and precise diagnosis of ASFV genes. As shown in Figure 1, biotin-ssDNA-azide and DBCO modified alkaline phosphatase (ALP) synthesized biotin-ssDNA-ALP through click chemistry, and biotin-ssDNA-ALP combined with streptavidin modified magnetic beads (SA-MB) to obtain the MB-ssDNA-ALP reporting system. The ASFV gene sequence was amplified with RPA to obtain amplicons. The amplicons bound to the Cas12a-crRNA complex, activated the trans-cleavage activity of Cas12a, cleaved ssDNA on MB-ssDNA-ALP, and released ALP; ALP catalyzed the hydrolysis of pNPP to produce pNP, which caused a change in colorimetric signal. Then, quantum dots were added as fluorescence reporters to realize colorimetric and fluorescence dual-mode signal output.
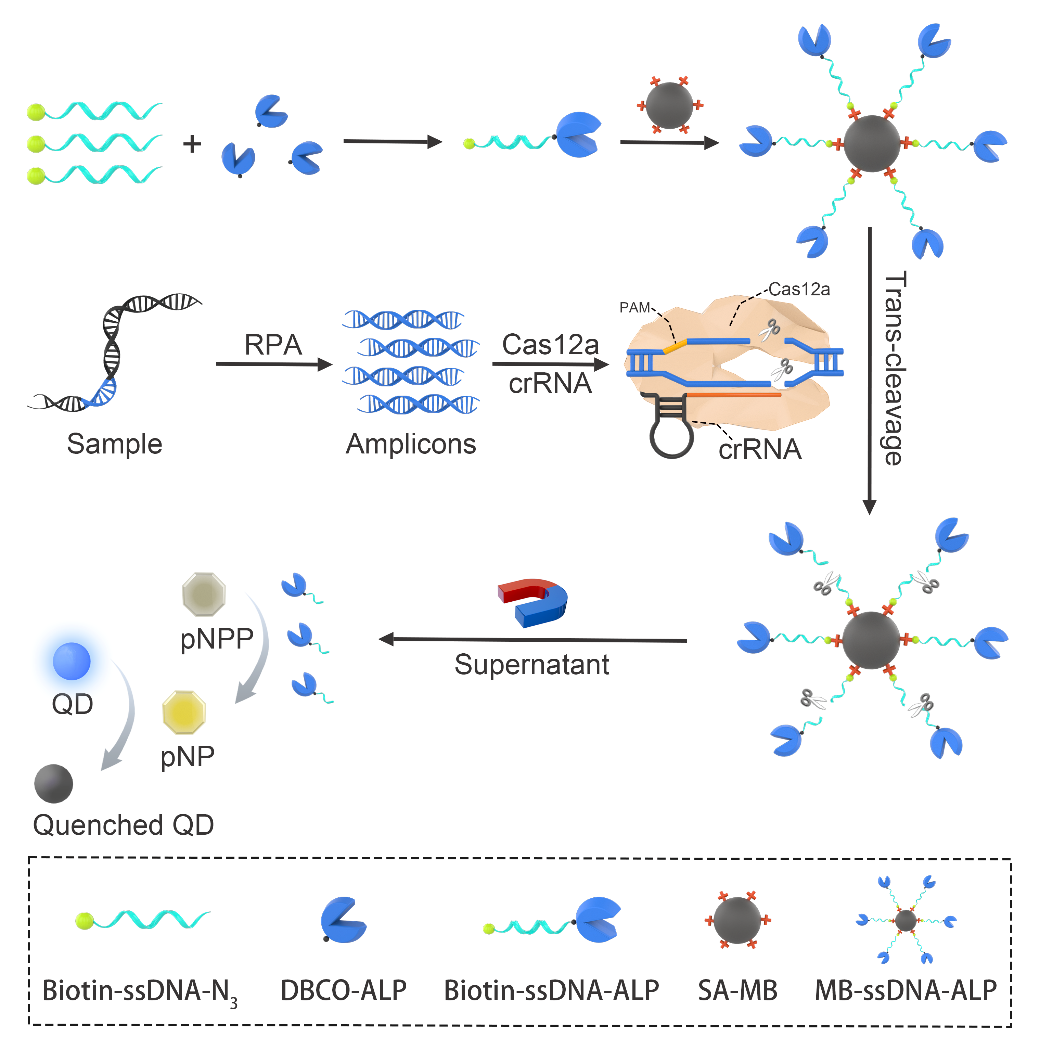
Figure 1 Schematic diagram of probe construction and ASFV detection. biotin-ssDNA-azide and DBCO-ALP synthesized biotin-ssDNA-ALP through click chemistry, and biotin-ssDNA-ALP was coupled with SA-MB to obtain MB-ssDNA-ALP reporter. RPA amplified the ASFV gene, the amplicon activated the Cas12a-crRNA complex, Cas12a non-specifically cleaved the biotin-ssDNA-ALP reporter, and ALP was released; ALP hydrolyzed pNPP to produce yellow pNP. After adding blue QDs, the fluorescence of blue QDs was quenched under the inner filter effect.
In this study, the team first synthesized and characterized biotin-ssDNA-ALP, and verified the coupling between biotin-ssDNA-N3 and DBCO modified ALP, as well as the synthesis of the MB-ssDNA-ALP probe. They then investigated the ability of RPA to amplify the ASFV gene. They designed three pairs of RPA primers targeting the highly conserved ASFV B646L gene, and verified that all three pairs of primers could efficiently amplify the ASFV gene by agarose gel electrophoresis experiments. Next, they verified the ability of CRISPR/Cas12a to target and cleave the ASFV gene. They designed crRNA targeting three RPA amplicon sequences, and verified that Cas12a was activated and cleaved the RPA amplicon using agarose gel electrophoresis. Finally, in conjunction with the synthesized MB-ssDNA-ALP probe, RPA-CRISPR/Cas12a system, and blue CdZnSe QDs, they achieved precise colorimetric - fluorescence dual-mode detection of the ASFV gene with a sensitivity of 20 copies/mL and visual detection of 104 copies/mL (Figure 2A-2D). The application of magnetic separation technology could effectively reduce background signals and achieve high SNR detection. By comparison, it was found that the detection method developed in this study had higher sensitivity than qPCR and good specificity. The probe did not cross-react with other swine disease related viruses. At the same time, they also explored the detectability of this method in real samples and analyzed 12 real samples of swine blood. The results showed that the detection method had 100% accuracy (Figure 2E and 2F). By combining the high sensitivity of fluorescence method and the legibility of colorimetry, this method has good sensitivity, portability, and specificity, providing an effective tool for rapid and accurate diagnosis of viral infection.

Figure 2 Colorimetric - fluorescence dual-mode detection of ASFV gene. (A) UV spectrum of pNP at different concentrations of the ASFV gene. (B) The linear relationship between absorbance at 400 nm and the logarithmic value of ASFV gene concentration. (C) Fluorescence spectrum of CdZnSe QDs at different concentrations of the ASFV gene. (D) The linear relationship between the change in fluorescence intensity and the logarithmic value of ASFV gene concentration. The analysis results of colorimetric (E) and fluorescence (F) detection methods applied to clinical samples. The visual detection results are all listed above the pictures.
Mao Guobin, an associate researcher of the Institute of Synthetic Biology of Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, and Luo Xing, a master degree candidate jointly trained by Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, and Huazhong Agricultural University, are the parallel first authors of the paper. Researcher Ma Yingxin, associate researcher Mao Guobin of the Institute of Synthetic Biology of Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, and Dr. Yin of Hubei University are the co-corresponding authors of the paper. Professor He Jin and associate professor Wang Xun of Huazhong Agricultural University participated in the study, and professor Kong Jilie of Fudan University provided sample support. The project was supported by several programs, such as the National Natural Science Foundation, and programs of the Ministry of Science and Technology, the Chinese Academy of Sciences, Guangdong Province, Shenzhen City and the Shenzhen Institute of Synthetic Biology.
supplementary cover story
Construction of ultra-highly sensitive CRISPR/Cas12a molecular diagnostic sensor for rapid screening of African swine fever virus infection
Introduction to PI and the research group:
Dr. Ma Yingxin, a researcher of Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, doctoral supervisor, winner of the Outstanding Youth Program of the National Natural Science Foundation, and Young Chief Scientist of the National Key Research and Development Program. In the past five years, he has presided over 1 Outstanding Youth Program of the National Natural Science Foundation, 1 Youth Program of the Key Research and Development Program of the Ministry of Science and Technology, 1 Youth Program of the National Natural Science Foundation, 1 Guangdong Outstanding Youth Foundation Program, 1 Shenzhen Excellent Scientific and Technological Innovation Talent Training Program, 1 Excellent Youth Program of Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, and 1 General Program of the Postdoctoral Science Foundation, and has been approved with talent programs such as Youth Innovation Promotion Association of the Chinese Academy of Sciences, Shenzhen High-level Talents, and Postdoctoral Innovative Talents Support Program.
He has published more than 30 papers as the first/corresponding author in internationally well-known journals such as J. Am. Chem. Soc., Nat. Commun., ACS Nano, Sci China Life Sci, Anal. Chem., ACS Appl. Mater. Interfaces, and Nano Res.
The research group is constantly recruiting postdoctoral fellows from:
1) those with research background in microbiology, bacteriophagology, synthetic biology, molecular biology, analytical chemistry, biomedicine;
2) those with work experience in molecular virology, bacteriophagology, synthetic biology, molecular biology, and development of biosensing technologies
3) Professionals with interdisciplinary backgrounds such as nanosynthetic biology
Interested applicants may send your resume (in PDF) to yx.ma1@siat.ac.cn. The subject line of the resume and email should be “Position applied - School Name - Major - Name”.